Here we are going to dock three ligands with a macromolecule. Select the hsg1 folder or the hsg1pdbqt file from the Macromolecules folder.

Tutorial Of Pyrx Software For Virtual Screening Youtube
Part 1 of the tutorial using AutoDock but Part 2 uses Vina.
. Specific steps for using PyRx as well as considerations for data preparation docking and data analysis are also. Cahill May 2015 2. Previously we provided an article on the installation of Pyrx in Ubuntu.
Ensure that you have Python 26 or newer and MS Visual C 2008 RP installed prior to installing PyRx. The tutorial will help viewers to use PyRx software for virtual screening of ligands towards potential drug targets. Download PyRx - Virtual Screening Tool for free.
PyRx enables Medicinal Chemists to run Virtual Screening from any platform and helps users in. Virtual Screening software for Computational Drug Discovery. You can dock multiple ligands.
The notes below give extra information or corrections on each step of the tutorial. Pyrx is a bioinformatics tool to perform virtual screening 1. Luhur Septiadi 15620102 Mata Kuliah Peminatan Bioinformatika Tutorial Ligand-Protein Docking menggunakan PyrX Virtual Screening Tools 1.
PyRx is a GUI for setting up and analysing AutoDock and Vina dockings - Relatively new - easy to use - works well. In this article we are going to perform protein-ligand docking using Pyrx. I have all molecules in a sdf format inside a.
A Beginners Manual for PyRx Drexel University John P. For virtual screening of. I am looking to do virtual screening of 2000 ligands to my target molecule.
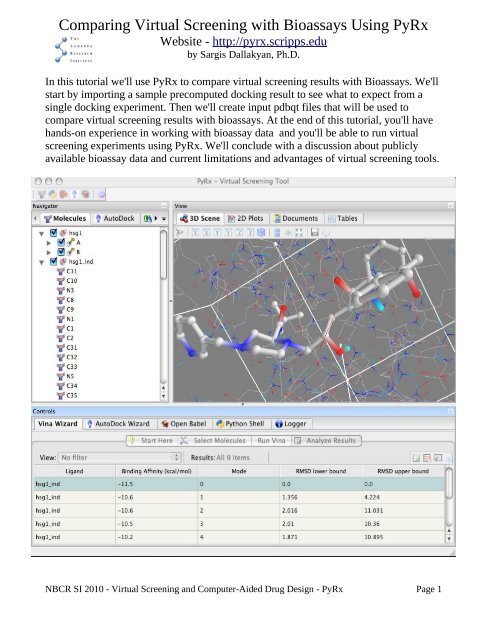
Make sure you are using the correct wizard in. Here we will dock one ligand only for demonstration. Compare virtual screening results with bioassays.
Hey all I am using the PyRx version 8 and earlier free version. Retrieve Data Sampel Protein Target dan Ligand Senyawa Aktif a. Pyrx 1 is a virtual screening software that allows the docking of multiple ligands with a target protein.
A Beginners Manual for PyRx 1. At the end of this tutorial youll have hands-on experience in working with bioassay data and youll be able to run virtual screening experiments using PyRx. We previously provided an article on performing simple protein-ligand site-specific docking using Pyrx.
PyRx is a Virtual Screening software for Computational Drug Discovery that can be used to screen libraries of compounds against potential drug targets. Click Forward to go to the next exercise. And carry out a virtual screen.
Move the spherical handles of. Sebelum melakukan Docking perlu di ambil data terlebih dahulu. This chapter describes how to perform small-molecule virtual screening by docking with PyRx which is open-source software with an intuitive user interface that runs on all major operating systems Linux Windows and Mac OS.
In this article we are going to perform virtual screening using Pyrx. Well conclude with a discussion about publicly available bioassay data and current limitations and advantages of virtual screening tools. 14 Running AutoGrid In the 3D Scene you will see a cube with a spherical handle on each face and one handle in the center.
13 Selecting Molecules Helpful instructions are on the screen Select indpdbqt from the Ligands folder.

Pyrx Virtual Screening Tool For Drug Discovery Youtube

Virtual Screening By Pyrx And Analysis Of Result By Ds Visualizer Youtube

A Screenshot Of Pyrx Virtual Screening Tool The Table On The Left Download Scientific Diagram

Pyrx Virtual Screening Tool Facebook
How To Perform Virtual Screening Using Pyrx Bioinformatics Review

0 comments
Post a Comment